Abstract
Aptamers are oligonucleotides or peptides with unique binding properties for specific target molecules,
and they have shown great potential in diagnostics, therapeutics, and bio-sensing. However, the current
in vitro SELEX-based method for discovering new target-selective aptamers is challenging, time-consuming,
and often unsuccessful in finding high-affinity aptamers. Recently, in silico methods have gained immense
attention. However, since labeled interaction-pair data collection is expensive and needs highly trained
specialists, available data is sparse. Further, since acquiring positive-class samples is even more
challenging, available datasets showcase high-class imbalance. This makes designing deep learning models
incredibly challenging, as they require a sufficiently large training set and are biased towards the dominant
class. Additionally, current models cannot be updated in real-time, and end-to-end re-training is necessary
for each new aptamer-target interaction pair discovery. The present work is the first to address both these
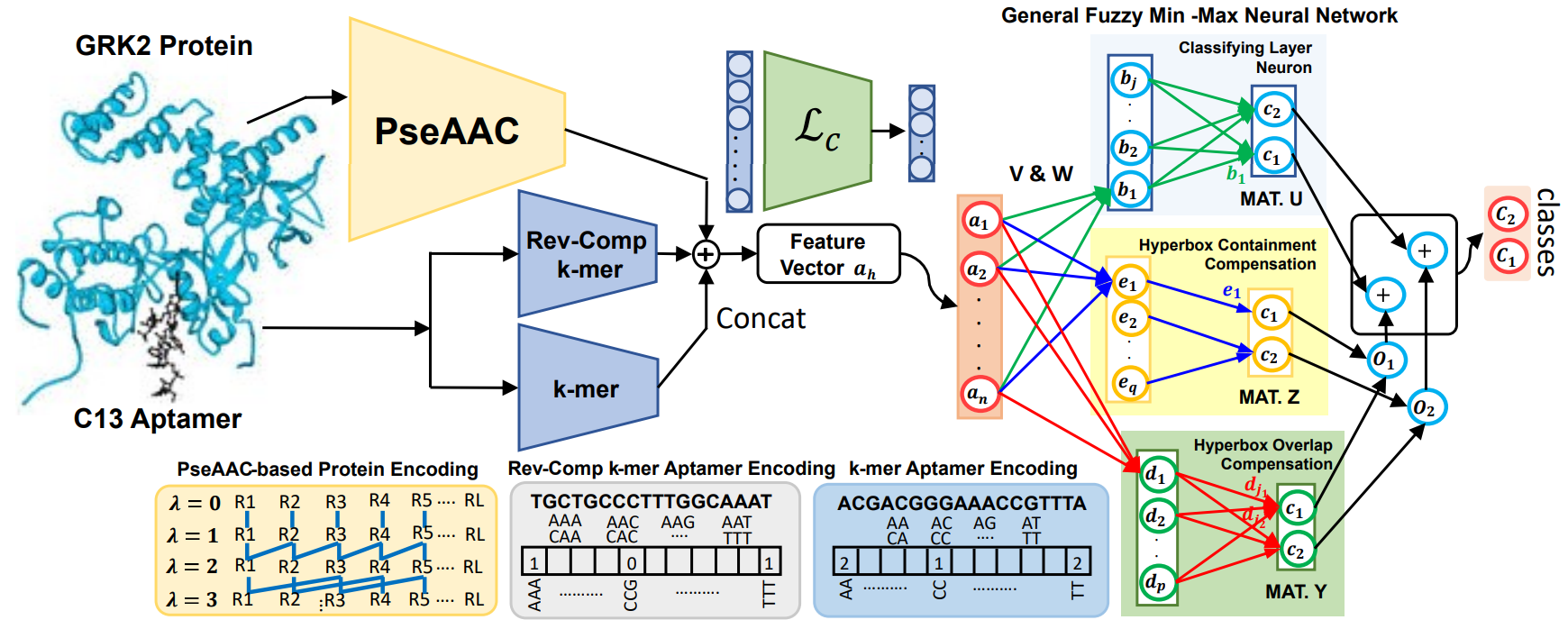
challenges. We present cAPTured, a novel fuzzy continual learning method for predicting aptamer-target protein
interaction pairs in a continual learning environment. cAPTured continually updates its learned feature space
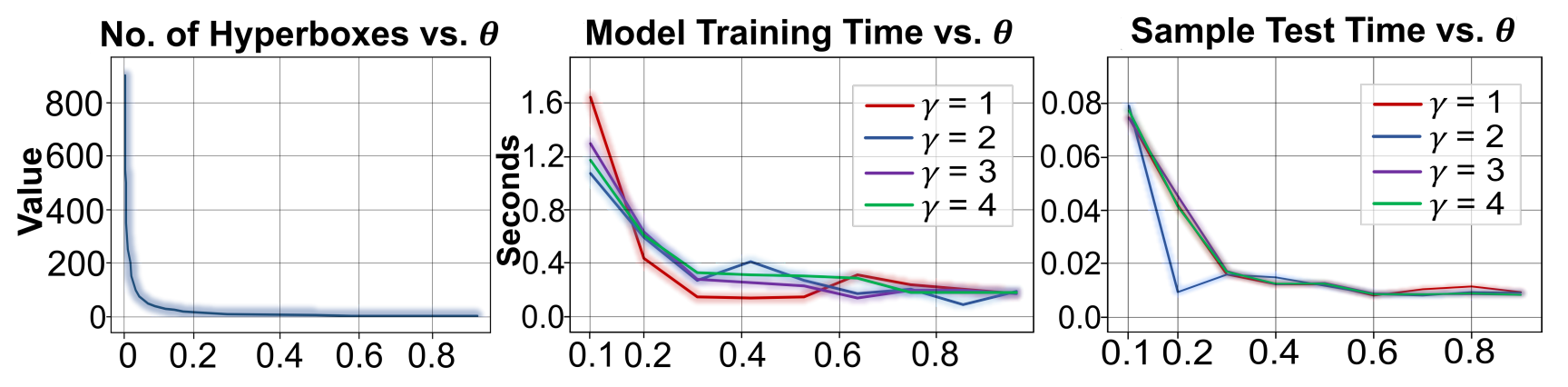
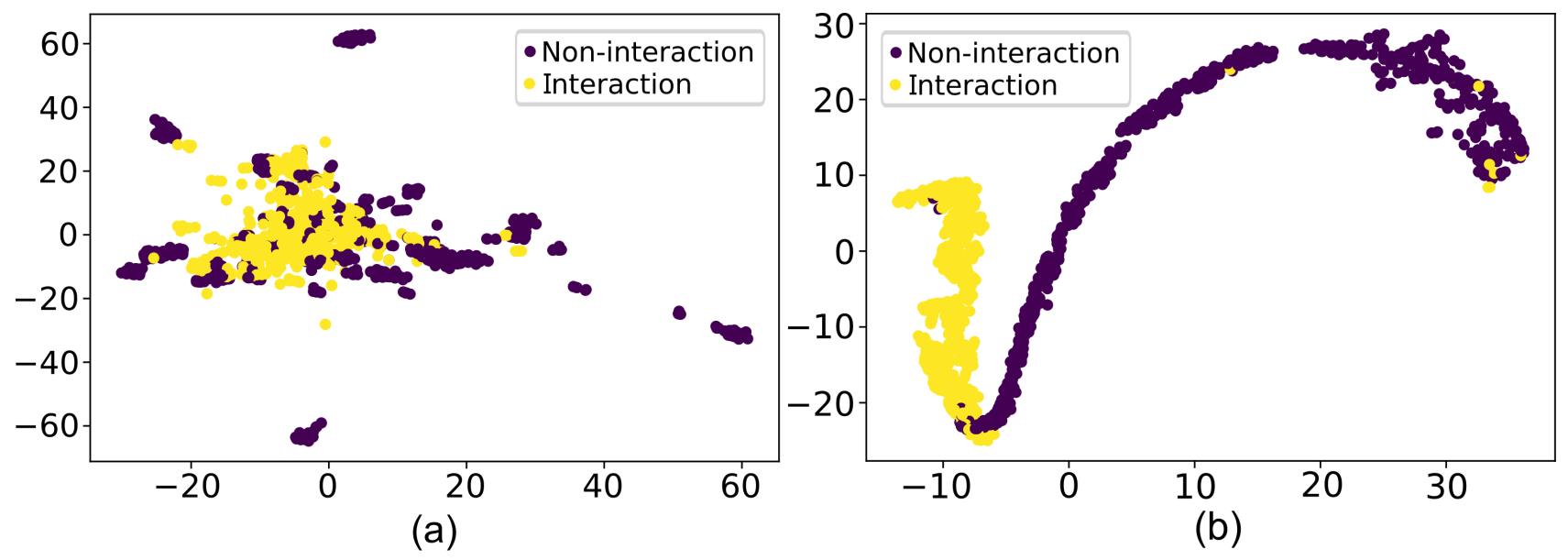
on a non-stationary interaction-pair data stream. We performed extensive evaluation studies and experiments
to establish the effectiveness of the proposed approach. cAPTured outperforms existing methods on the
benchmark dataset by a significant margin.